Docker container for locator NN. See: https://github.com/kr-colab/locator.git
Download the Dockerfile
(you should also download Docker Desktop)
Create an image with the Dockerfile. Open terminal (or similar) and run the following:
docker build -t locator .
You must be inside of the directory containing the Dockerfile for this to work.
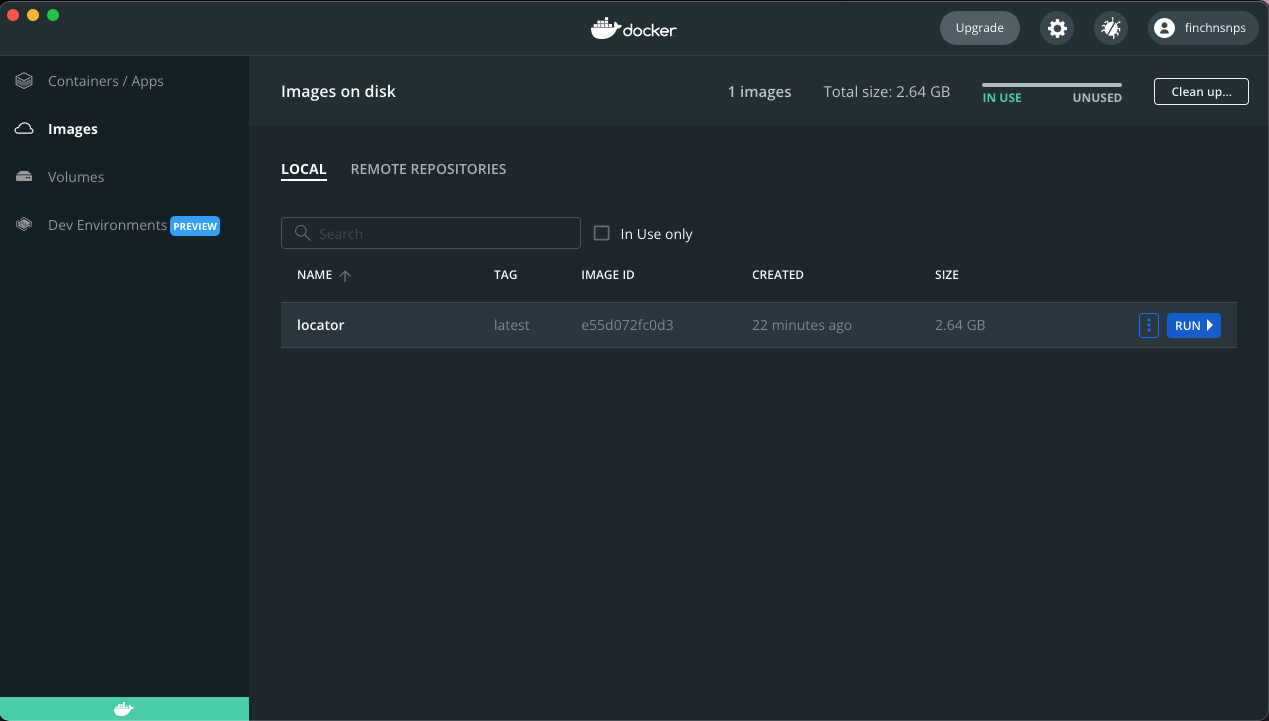
See image in you Docker Desktop app.
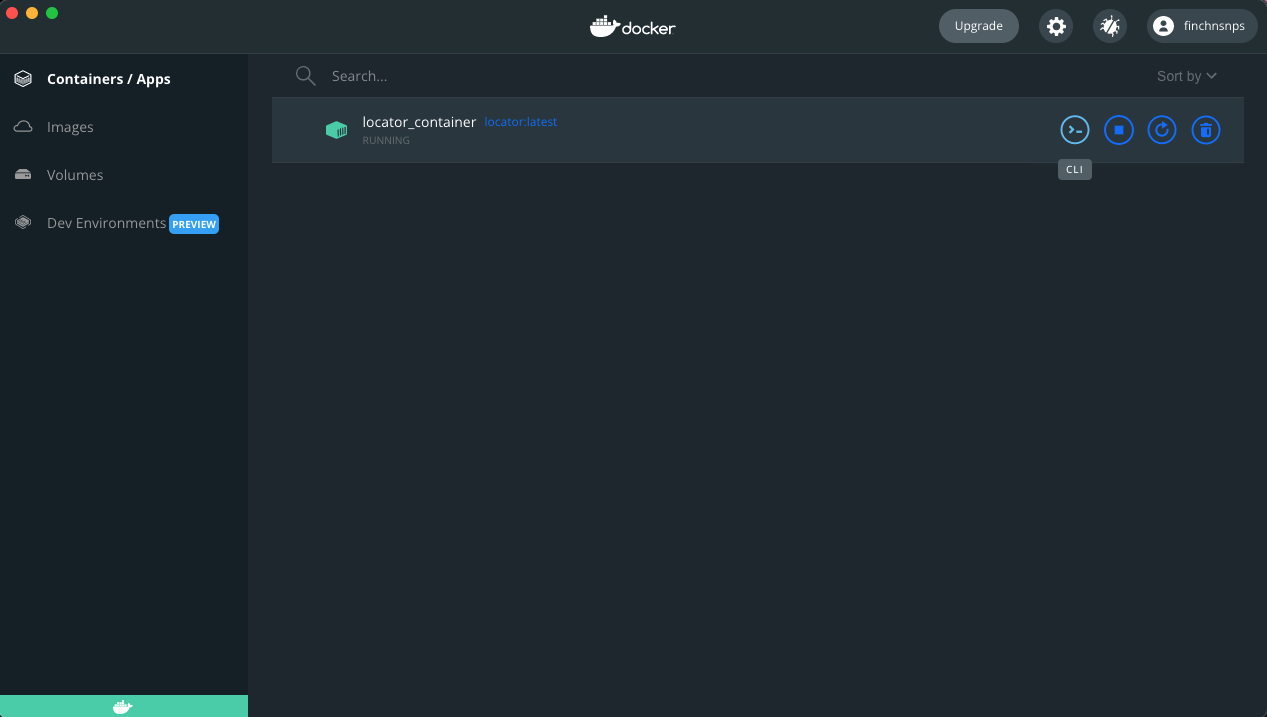
Click the RUN! button on the right side; name the container (optional).
Click the >_ button which will open the container in terminal.
Find the scripts directory.
ls scripts
Load module first if necessary.
module load singularity
Download docker image to be run with singluarity.
singularity pull locator.sif docker://finchnsnps/locator:locator_v2
Open container as a shell with singularity.
singularity shell --cleanenv locator.sif
Find locator scripts directory under /
ls /locator/scripts
See https://github.com/kr-colab/locator.git for details about test data, output files, parameters, visualization, and interpretation.
Make output directory
mkdir out/test
Use test data to test the container
In docker:
python scripts/locator.py --vcf data/test_genotypes.vcf.gz --sample_data data/test_sample_data.txt --out out/test/test
In singularity
python /locator/scripts/locator.py --vcf /locator/data/test_genotypes.vcf.gz --sample_data /locator/data/test_sample_data.txt --out out/test/test